Note
Click here to download the full example code or run this example in your browser via Binder
Using a Surrogate Tree Explainer¶
This example illustrates how to use a tabular surrogate tree-based explainer to explain a prediction.
This example shows how to use the tabular surrogate tree-based explainer
implementation –
fatf.transparency.predictions.surrogate_explainers.TabularBlimeyTree
– to explain a prediction of a black-box probabilistic model.
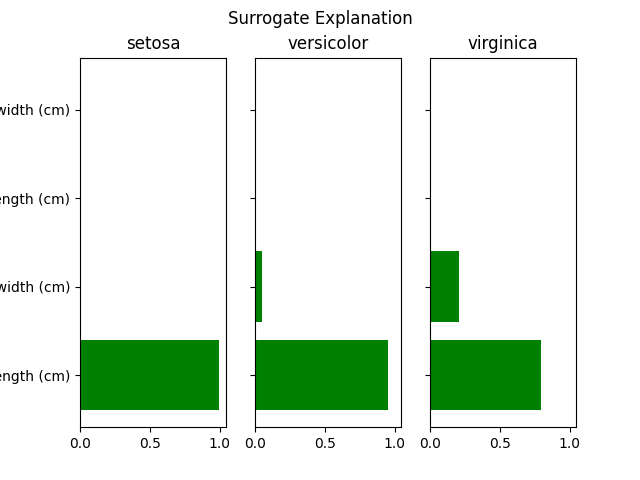
Out:
{'setosa': {'petal length (cm)': 0.9947582385676036,
'petal width (cm)': 0.0,
'sepal length (cm)': 0.0,
'sepal width (cm)': 0.00524176143239636},
'versicolor': {'petal length (cm)': 0.9496406928798405,
'petal width (cm)': 0.05035930712015947,
'sepal length (cm)': 0.0,
'sepal width (cm)': 0.0},
'virginica': {'petal length (cm)': 0.7962708998702057,
'petal width (cm)': 0.20372910012979428,
'sepal length (cm)': 0.0,
'sepal width (cm)': 0.0}}
# Author: Kacper Sokol <k.sokol@bristol.ac.uk>
# License: new BSD
from pprint import pprint
import fatf
import fatf.utils.data.datasets as fatf_datasets
import fatf.utils.models as fatf_models
import fatf.transparency.predictions.surrogate_explainers as fatf_surrogates
import fatf.vis.lime as fatf_vis_lime
print(__doc__)
# Fix random seed
fatf.setup_random_seed(42)
# Load data
iris_data_dict = fatf_datasets.load_iris()
iris_X = iris_data_dict['data']
iris_y = iris_data_dict['target']
iris_feature_names = iris_data_dict['feature_names'].tolist()
iris_class_names = iris_data_dict['target_names'].tolist()
# Train a model
clf = fatf_models.KNN()
clf.fit(iris_X, iris_y)
# Create a LIME explainer
tree = fatf_surrogates.TabularBlimeyTree(
iris_X,
clf,
feature_names=iris_feature_names,
class_names=iris_class_names)
# Choose an index of the instance to be explained
index_to_explain = 42
# Explain an instance
tree_explanation = tree.explain_instance(
iris_X[index_to_explain, :], samples_number=500)
# Display the textual explanation
pprint(tree_explanation)
# Plot the explanation
fatf_vis_lime.plot_lime(tree_explanation)
Total running time of the script: ( 0 minutes 2.179 seconds)

